Python and its various libraries have some cool built-in functions. However, as an earth system scientist, I feel it sorely lacks a function to compare N-dimension datasets. Much of earth science data is of (time, lat, lon) nature, and as far as I am aware, there is no library that lets you compute basic statistics such as covariance, correlation, and regression between two multi-dimentional datasets along a given axis.
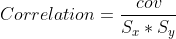
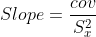
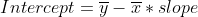
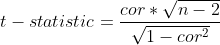

I first used to apply brute force and do these computations using available functions (e.g. scipy.stats.linregress()) after looping over lat and lon dimensions. i.e. double looping. As the data resolutions increased, I tried to parallelize the looping. However, I soon realized that vectorizing these statistics would not be that difficult, and depending on data size, more than 1000s times faster! Hence I wrote a function that would compute same statistics as stats.linregress(), but with multi-dimensional data in mind. Also, as several geophysics variables show lagged relationships, I have incorporated that functionality as well. Also, the two datasets, or one of them could also be a 1D time series. This way, the function is more robust, and useful to compare a single time series such as an ENSO index to a spatial dataset.
Let me walk you through the function. Like my previous Python tutorials, I have used GPCP data. You can download it from here: https://www.esrl.noaa.gov/psd/data/gridded/data.gpcp.html. Also, since annual cycle and trend affect correlation, I usually remove them, as demonstrated. Depending on what you want, you may or may not want to do this. It's certainly not required for the function to run! Also, I am not a computing whiz, so please let me know if there are more efficient ways to represent this.
Let me walk you through the function. Like my previous Python tutorials, I have used GPCP data. You can download it from here: https://www.esrl.noaa.gov/psd/data/gridded/data.gpcp.html. Also, since annual cycle and trend affect correlation, I usually remove them, as demonstrated. Depending on what you want, you may or may not want to do this. It's certainly not required for the function to run! Also, I am not a computing whiz, so please let me know if there are more efficient ways to represent this.
In [1]:
%matplotlib inline
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats, signal #Required for detrending data and computing regression
In [2]:
"""Load data"""
ds = xr.open_dataset('../../../InputData/Precipitation/GPCP/GPCP2.3/precip.mon.mean.nc')
data = ds.precip
#Plot global mean time series
plt.figure(figsize=(15, 5))
plt.plot(data.time, data.mean(axis=(1,2)), label = 'original')
plt.ylabel(data.units)
print data.dims, data.shape
"""Remove seasonality as it would influence the correlation:
Take a 12-month moving average;
The average will affect all values in the first and last 6 months, so remove them.
"""
data = data.rolling(time= 12, center=True).mean()[6:-6,...]
print data.shape
plt.plot(data.time, data.mean(axis=(1,2)), label = '12-mo ma')
mean = data.mean(axis=0)
"""Detrend data as it would influence the correlation"""
data = xr.DataArray(signal.detrend(data, axis=0), dims=data.dims, coords=data.coords) + mean
plt.plot(data.time, data.mean(axis=(1,2)), label = '12-mo ma & detrended')
plt.legend(loc = 'upper left', ncol = 3).draw_frame(False)
plt.title('Mean Global Precipitation')
Out[2]:
In [3]:
"""For Testing, get time series from two random grid points"""
ts1 = data[:,36,116]
ts2 = data[:,39,52]
ts1.plot()
ts2.plot()
Out[3]:
As you can see, the time series have some correlation, but not much.
Here's the function, commented for clarity:
Here's the function, commented for clarity:
In [4]:
def lag_linregress_3D(x, y, lagx=0, lagy=0):
"""
Input: Two xr.Datarrays of any dimensions with the first dim being time.
Thus the input data could be a 1D time series, or for example, have three dimensions (time,lat,lon).
Datasets can be provied in any order, but note that the regression slope and intercept will be calculated
for y with respect to x.
Output: Covariance, correlation, regression slope and intercept, p-value, and standard error on regression
between the two datasets along their aligned time dimension.
Lag values can be assigned to either of the data, with lagx shifting x, and lagy shifting y, with the specified lag amount.
"""
#1. Ensure that the data are properly alinged to each other.
x,y = xr.align(x,y)
#2. Add lag information if any, and shift the data accordingly
if lagx!=0:
#If x lags y by 1, x must be shifted 1 step backwards.
#But as the 'zero-th' value is nonexistant, xr assigns it as invalid (nan). Hence it needs to be dropped
x = x.shift(time = -lagx).dropna(dim='time')
#Next important step is to re-align the two datasets so that y adjusts to the changed coordinates of x
x,y = xr.align(x,y)
if lagy!=0:
y = y.shift(time = -lagy).dropna(dim='time')
x,y = xr.align(x,y)
#3. Compute data length, mean and standard deviation along time axis for further use:
n = x.shape[0]
xmean = x.mean(axis=0)
ymean = y.mean(axis=0)
xstd = x.std(axis=0)
ystd = y.std(axis=0)
#4. Compute covariance along time axis
cov = np.sum((x - xmean)*(y - ymean), axis=0)/(n)
#5. Compute correlation along time axis
cor = cov/(xstd*ystd)
#6. Compute regression slope and intercept:
slope = cov/(xstd**2)
intercept = ymean - xmean*slope
#7. Compute P-value and standard error
#Compute t-statistics
tstats = cor*np.sqrt(n-2)/np.sqrt(1-cor**2)
stderr = slope/tstats
from scipy.stats import t
pval = t.sf(tstats, n-2)*2
pval = xr.DataArray(pval, dims=cor.dims, coords=cor.coords)
return cov,cor,slope,intercept,pval,stderr
Equations in the function are as follows. Note that the mean and standard deviations mentioned here should be calculated along time axis only.
(y-\overline{y})}{n})
P-value is computed from t-statistic. In python, this can be done by scipy.stats.t.sf(tstats,n-2)
(For a two-tailed test, multiply this value by 2)
(For a two-tailed test, multiply this value by 2)
Now lets call the function. To test it, lets regress ts1 with our 3D data. Thus, from the result, [39,52] will be same as comparing the two time series ts1 and ts2. This way we can compare the above function directly with stats.linregress(). Finally, lets also compare ts1 and ts2 as well with our new function, to demonstrate data could be just 1-dim time series as well (but of course one could just use linregress() for that).
In [5]:
cov,cor,slope,intercept,pval,stderr = lag_linregress_3D(x=ts1,y=data)
print 'x = 1-dim, y = 3-dim:\n', cor[39,52].data, pval[39,52].data, slope[39,52].data, intercept[39,52].data, stderr[39,52].data
ans = stats.linregress(ts1,ts2)
print 'stats.linregress() output:\n',ans.rvalue, ans.pvalue, ans.slope, ans.intercept, ans.stderr
cov,cor,slope,intercept,pval,stderr = lag_linregress_3D(ts1,ts2)
print 'x = 1-dim, y = 1-dim:\n',cor.data,pval.data,slope.data,intercept.data,stderr.data
Cool.
Now let's check the lag relationship by providing lag=1 for ts1:
Now let's check the lag relationship by providing lag=1 for ts1:
In [6]:
cov,cor,slope,intercept,pval,stderr = lag_linregress_3D(x=ts1,y=data, lagx=1)
print cor[39,52].data, pval[39,52].data, slope[39,52].data, intercept[39,52].data, stderr[39,52].data
"""To compare, we will have to remove the first value from ts1, and last from ts2"""
ans = stats.linregress(ts1[1:],ts2[:-1])
print ans.rvalue, ans.pvalue, ans.slope, ans.intercept, ans.stderr
Thats all folks!
4 comments:
Nice Post. Thanks for sharing...
thank you for sharing
toko obat kuat pria
obat kuat jakarta
obat kuat viagra
obat kuat asli
viagra jakarta
viagra usa
toko viagra original
thanks, this is really helpful!
Hi Hrishi,
Thanks for an amazing function!
I just ran into a small issue for some time series where the t-score were negative and the p-value did not match with the ones calculated with other linear regression packages.
I think it is the absolute value of the t-score that should be given to the Student's t survival function:
pval = t.sf(abs(tstats), n-2)*2
instead of
pval = t.sf(tstats, n-2)*2
Thanks again for your work!
Post a Comment